The innovation field curATech combines curAIdent and curATreat by creating bioinformatic foundations to simplify the identification of biomarkers with the help of artificial intelligence and to develop and understand innovative technologies for the analysis of biodata on vascular functions and structures.
P10 curAIscid - curATime AI science and development
curAIscid aims to develop widely applicable solutions to cross-project AI problems. The following four topics have been identified as essential in advance:
Small data: The problem of small datasets is to be addressed through approaches such as transfer learning, data augmentation as well as knowledge-intensive machine learning.
Explainability: AI-based predictions may be due to purely associative rather than causal factors, highlighting the need to combine both explainable AI and formal causal inference methods.
Representation Learning and Phenotyping: The use of e.g. autoencoder-assisted dimensionality reduction can reduce complexity and noise in multidimensional data.
Privacy and Fairness: In order to ensure data protection and confidentiality, the project will investigate concepts of differential privacy.
 Prof. Dr. Sebastian Vollmer
Prof. Dr. Sebastian Vollmer
Coordinator curAIscid
Head of the Research Area Data Science and its Applications, German Research Center for Artificial Intelligence GmbH
[javascript protected email address]
P11 curAIvasc - AI-based analysis of vascular imaging for optimized optimised multidimensional information assessment
curAIvasc aims to develop innovative technologies and pipelines for the analysis of multi-dimensional biodata on vascular function and structure and to transfer them into patient-oriented research.
In close collaboration, the German Research Centre for Artificial Intelligence and the University Medical Center Mainz will analyse vascular imaging data from the large-scale y Gutenberg Health Study and disease-specific cohorts. Standardised pre-processing steps, harmonisation processes and quality controls create the basis for an AI-based cross-cohort exploration of the data, leading to the development ofa modern Deep Learning-based multi-dimensional pipeline for image-based prediction of individual disease progression and personalised risk assessment. The identification of the most influential variables will help to transfer new biologically based influencing factors for risk assessment into medical research and to create a better understanding of the molecular processes of atherothrombosis.
 Univ.-Prof. Dr. Prof. h.c. Andreas Dengel
Univ.-Prof. Dr. Prof. h.c. Andreas Dengel
Coordinator curAIvasc
Managing Director German Research Center for Artificial Intelligence GmbH
[javascript protected email address]
P12 curAIheart - Artificial intelligence based evaluation of echocardiographic image data under consideration of high-dimensional clinical data
Echocardiography is a clinically established procedure for determining structure and functional parameters of the heart for classification and risk assessment of cardiovascular diseases. This is a laborious process, which in the clinic has so far been based on the visual evaluation of recorded video loops by clinical staff. Current methods still rely on the selection of defined image regions by medical staff and depend heavily on expertise and experience.
In curAIheart, innovative machine learning methods will be used to automate deep analysis of echocardiographic images and digitally recorded image information. The analysis of the recordings will incorporate the image loops themselves as well as data from molecular biology that will enable a deeper analysis. The pipeline to be developed will improve risk assessment and early detection of cardiovascular disease and will allow integration with other diagnostic data.
 Univ.-Prof. Dr. Stefan Kramer
Univ.-Prof. Dr. Stefan Kramer
Coordinator curAIheart
Institute for Informatics, Johannes Gutenberg University Mainz
[javascript protected email address]
P13 curAIsig - High-dimensional robust signal processing
curAIsig develops innovative technologies in systems-oriented biomedical research. In today's clinical practice, analytical methods are used that can lead to incorrect interpretations of results, especially in the case of data outliers and small sample sizes placed in relation to dimensionality. We will therefore develop and apply robust signal processing and statistical learning methods to discover novel and reproducible biomarker signatures.
The first contribution of curAIsig deals with autonomic dysfunctions leading to the development of atherosclerotic diseases and their consequences. New methods will be developed to extract clinically robust biomarkers from heart rate variability measurements. Furthermore, novel learning methods will be explored to develop clinically interpretable models from high-dimensional multi-omics and resulting data. We will apply these to biodatabases of the University Medical Center Mainz and will evaluate them in the context of the pathophysiology of atherothrombosis.
 Prof. Dr.-Ing. Abdelhak Zoubir
Prof. Dr.-Ing. Abdelhak Zoubir
Coordinator curAIsig
Signal Processing Group, Technical University of Darmstadt
[javascript protected email address]
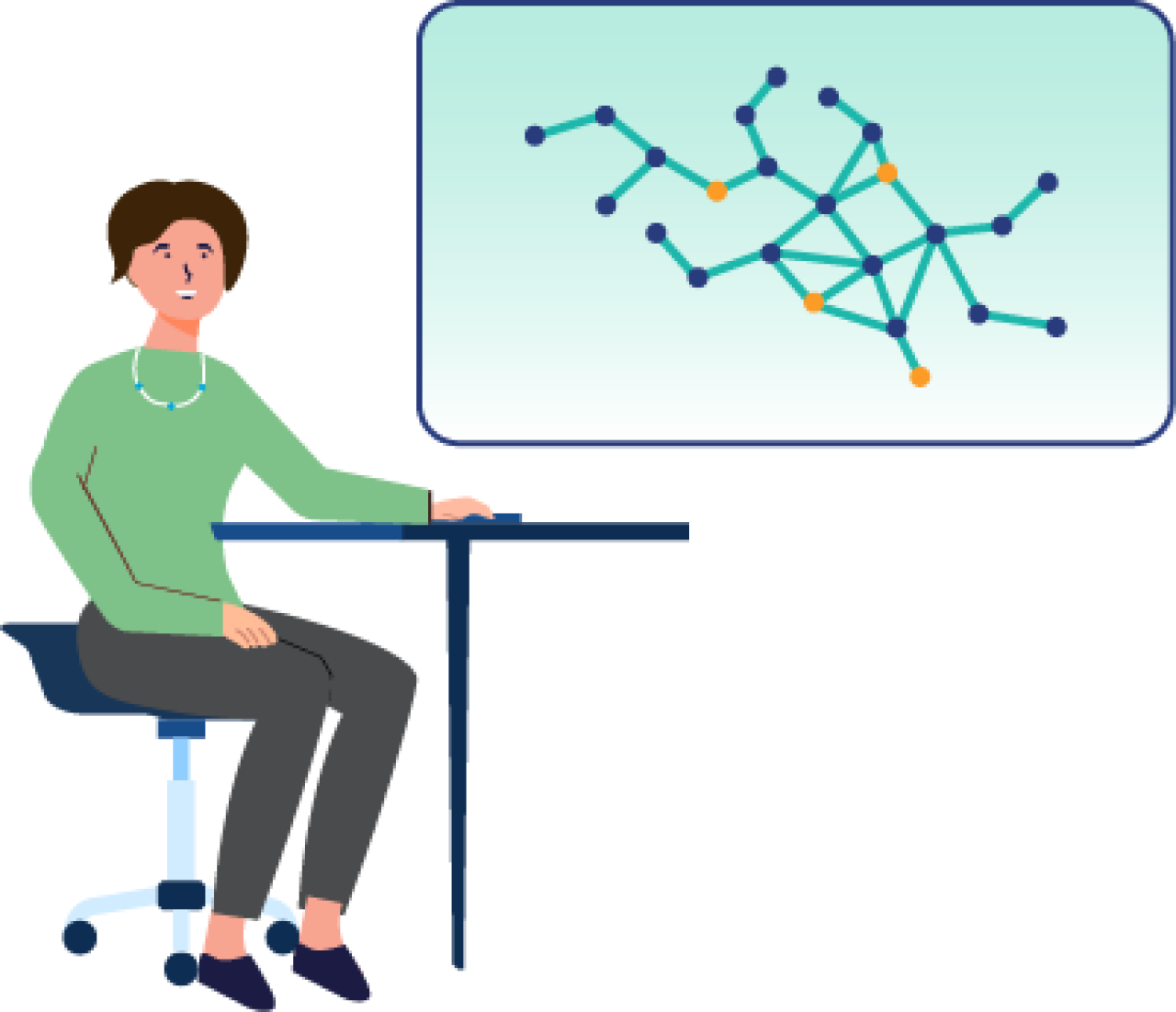
P14 curAIknow - Application of the life science knowledge graph Ontosight® to support the prediction and validation of AI-generated hypotheses on the basis of patient data and healthy controls
curAIknow deals with the development of knowledge-based machine learning methods based on a very large biomedical knowledge graph for use within the curATime cluster. An existing knowledge graph, Ontosight, covering almost all relevant public knowledge in the biosciences, will be combined with large-scale data from healthy individuals and patients for integrated analysis. Two directions will be pursued: On the one hand, the use of knowledge a priori in a machine learning algorithm. On the other hand, the use of knowledge a posteriori to retrospectively verify purely data-driven learned machine learning models in light of existing knowledge. Part of this task is the development of a so-called discovery agent that discovers or evaluates connections between two entities in the knowledge graph.
 Univ.-Prof. Dr. Stefan Kramer
Univ.-Prof. Dr. Stefan Kramer
Coordinator curAIknow
Institute for Informatics, Johannes Gutenberg University Mainz
[javascript protected email address]